From pink to blue to near-IR, protein stains come in a wide variety of colors. As protein biologists, these stains play a crucial role in our day-to-day lab work for routine protein analysis and western blot normalization. In fact, several studies have found that normalization by total protein is superior to normalizing with housekeeping proteins such as tubulin or actin.1–3 Yet each protein stain is unique, with inherent advantages and disadvantages that should be considered when interpreting results. In honor of our newly released protein prestain, VersaBlot™, we decided to give our readers a guide for the some of the most common protein stains which includes a close look at their origins, advantages and limitations.
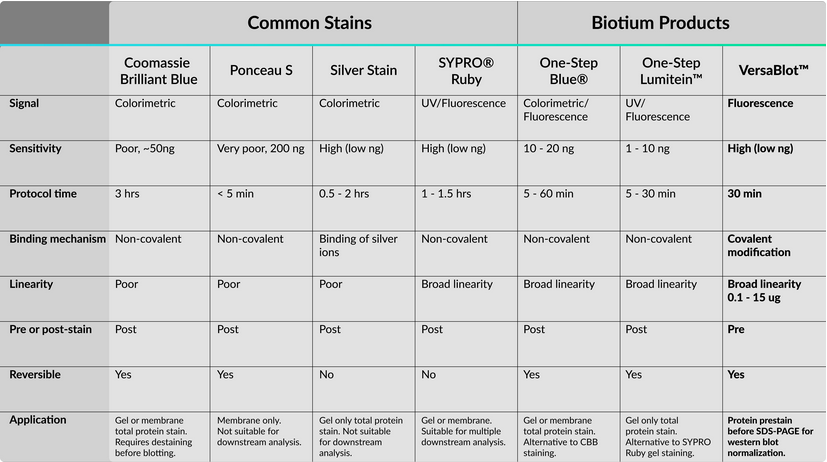
Here is a helpful table summarizing the protein stains discussed in this article and stains offered by Biotium.
The “Classic” Blue Coomassie
A guide for protein staining should rightfully begin with the most widely used protein stain, Coomassie Brilliant Blue (CBB). CBB was first used as an acid wool dye in the 19th century by a British dye manufacturer named Levinstein Ltd. Fast-forward to 1964, an Australian biochemist named Fazekas De St. Growth and colleagues discovered its application for staining proteins after gel electrophoresis.4 Since then, several versions of CBB have been developed, including the widely accepted “colloidal” G-250 formulation that’s more sensitive and has less background than the older R-250 form. CBB is widely popular in labs because the staining is cost-effective and easy to visualize. In addition, the staining protocol requires only a single prepared reagent and binds to proteins non-covalently through interactions with basic and aromatic residues. Despite these advantages, CBB lacks sensitivity relative to alternative staining methods (ie. silver stain, fluorescence) and has a narrow linear range. Moreover, membranes prestained with CBB must be completely destained or the dye may interfere with downstream analysis.
Advantages: Cost-effective and easy to visualize.
Disadvantages: Lacks sensitivity, small linear range, dye may interfere with downstream antibody binding and mass spectrometry analysis.
Use for: Gel or membrane when you need an inexpensive total protein stain. If you plan on western blotting, stain the membrane after transfer and before blocking.
Alternatives: Check out One-Step Blue® as a more convenient and versatile alternative to CBB staining.
Ponceau S, Pink Proteins on the Go
Ponceau, French for “poppy-colored”, describes a family of red Azo dyes used as food colorants and histology stains. What we know as Ponceau in our labs is the Ponceau S formulation, a reversible total protein stain for western blot membranes. Like CBB, Ponceau also binds to proteins non-covalently through ionic and hydrophobic interactions. Yet, unlike CBB’s longer staining protocol (at least 45 minutes plus destaining) Ponceau staining can be done in a few minutes and can be quickly destained with water. These conveniences allow Ponceau to be a quick method for visualizing total protein on western blots. However, where Ponceau gains in convenience it loses in sensitivity. Ponceau is one of the least sensitive staining techniques with a detection minimum of 200 ng.5 In addition, the stain is considerably weaker than CBB and may fade overtime, making normalization less reproducible between experiments.
Advantages: Cheap, quick and reversible.
Disadvantages: Poor sensitivity and reproducibility. Not recommended for normalization.
Use For: Membrane stain only as quick method to verify protein transfer efficiency for western blotting.
The Sensitive Silver Stain
Silver staining is considered the gold standard for sensitive protein detection methods. The technique was first developed for SDS-PAGE by Switzer et. al in 1979 which enabled sub-nanogram detection and 50-fold greater sensitivity relative to CBB staining.6 Interestingly, the mechanism behind this sensitivity relies on chemistry that’s similar to developing film in photography. Protein gels are impregnated with silver ions (Ag+) which bind to various amino acid functional groups. The silver is then reduced to silver metal, a process that is self-catalytic, allowing for ultrasensitive detection of protein bands. Unfortunately, this sensitivity is not without some serious drawbacks. The staining process can be very technical and has a narrow linear range, leading to very poor reproducibility. Moreover, the staining is irreversible and for gel use only and therefore not suited for downstream analysis such as mass spectrometry or western blotting.
Advantages: Cheap with excellent sensitivity.
Disadvantages: Time consuming, poor reproducibility and linearity, not compatible with downstream analysis.
Use for: Gel stain only when you need sub-nanogram sensitivity for total protein.
SYPRO® Ruby, A New Era
Despite how popular and inexpensive colorimetric stains are, drawbacks with sensitivity, linearity and compatibility with downstream analysis prompted development of more advanced protein detection methods. Enter SYPRO® Ruby, a ruthenium based fluorescent dye that was developed by Molecular Probes in 2000.7 SYPRO® Ruby staining allowed for silver stain sensitivity but with greater linear range and compatibility with downstream analysis. Additionally, the ease of use and high reproducibility of SYPRO® Ruby as a total protein stain provided a superior method for western blot normalization over traditional house-keeping proteins.1 Over a decade later, several other fluorescence-based protein stains have been developed including LI-COR REVERT™, Bio-Rad Stain-Free™ and Amersham™ Quickstain. One disadvantage for fluorescent stains is they are generally more expensive than colorimetric methods and require a fluorescence imager. However, as the research community gradually shifts toward fluorescence-based technologies, fluorescence imagers are becoming far more common and affordable.
Advantages: Convenient, sensitive, wide linear range and compatible with multiple downstream analysis.
Disadvantages: More expensive than colorimetric methods and requires a fluorescence imager.
Use for: There are SYPRO ® Ruby formulations specific for gel or membrane protein detection. Can be used for several applications including western blot normalization and mass spectrometry.
Alternatives: Check out One-Step Lumitein™ and One-Step Lumitein™ UV as a low cost and more convenient alternative to SYPRO ® Ruby.
VersaBlot™, Convenient, Highly-Linear and Reversible
Biotium has furthered the advancement of fluorescent protein detection with VersaBlot™, a highly linear, sensitive and reversible prestain. Unlike colorimetric or fluorescent protein stains where staining is done after running your gel, VersaBlot™ is added to your sample before electrophoresis and can be used for downstream western blot normalization. The VersaBlot™ Total Protein Normalization Kit comes with your choice of two near-IR CF® Dyes for emission in Odyssey® 700 or 800 channels.
Advantages: Convenient, sensitive with a wide linear range. Signal can be quenched for downstream multi-color near-IR detection.
Disadvantages: More expensive than colorimetric methods, requires a near-IR fluorescence imager.
Use for: A convenient method for western blot normalization and downstream multi-color near-IR protein detection.
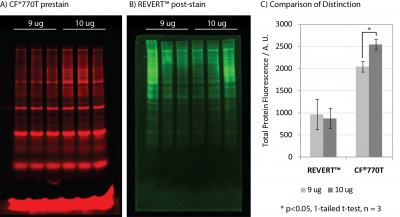
VersaBlot™ CF®770T prestain demonstrated superior signal-to-noise and sensitivity relative to LI-COR® REVERT™ kit
Discover our new VersaBlot™ Total Protein Normalization Kits.
Cited References
- Li, R. & Shen, Y. Life Sciences 92, 747–751 (2013).
- Hu, X. et al. Oncotarget 7, 66679–66688 (2016).
- Aldridge, G. M. et al. J. Neurosci. Methods 172, 250 (2008).
- Fazekas de St Groth, S. et al. Biochim. Biophys. Acta 71, 377–91 (1963).
- Goldman, A. et al. Curr. Protoc. Protein Sci. 2016, 10.8.1-10.8.11 (2016).
- Switzer, R. C. et al. Anal. Biochem. 98, 231–7 (1979).
- Berggren, K. et al. Electrophoresis 21, 2509–21 (2000).
- Chevallet, M. et al. Nat. Protoc. 1, 1852–1858 (2006).